Chapter 6: Smoothing
Why Smooth?
It is common to smooth the functional data, or replace the signal at each voxel with a weighted average of that voxel’s neighbors. This may seem strange at first - why would we want to make the images blurrier than they already are?
It is true that smoothing does decrease the spatial resolution of your functional data, and in general we don’t want less resolution. But there are benefits to smoothing as well, and these benefits can outweigh the drawbacks. For example, we know that fMRI data contain a lot of noise, and that the noise is frequently greater than the signal. By averaging over nearby voxels we can cancel out the noise and enhance the signal.

In this animation, two different smoothing kernels (4mm and 10mm) are applied to an fMRI scan. Notice that as we use larger smoothing kernels, the images become blurrier and the anatomical details become less distinct. Also note that, for the sake of simplicity, this animation uses a 2D slice of the brain to demonstrate this preprocessing step. In actual fMRI data, the kernel would be applied in all three dimensions.
How to Smooth in SPM
In the SPM GUI, click on the Smooth button and double-click on Images to Smooth. Select the warped functional images, and expand them to include all 146 frames for each run. (See the previous chapters for examples on how to use the Filter and Frames fields to select the images that you want.) Leave the other defaults as they are, and then click on the green Go button.
Note
The default FWHM of 8x8x8mm is probably too large for most studies; it may help boost the signal over larger cortical regions, but it is likely to dilute the signal of functionally smaller but more homogeneous regions. Once you have finished this series, you are encouraged to test different smoothing kernels to see how it affects the extent and the significance of your results.
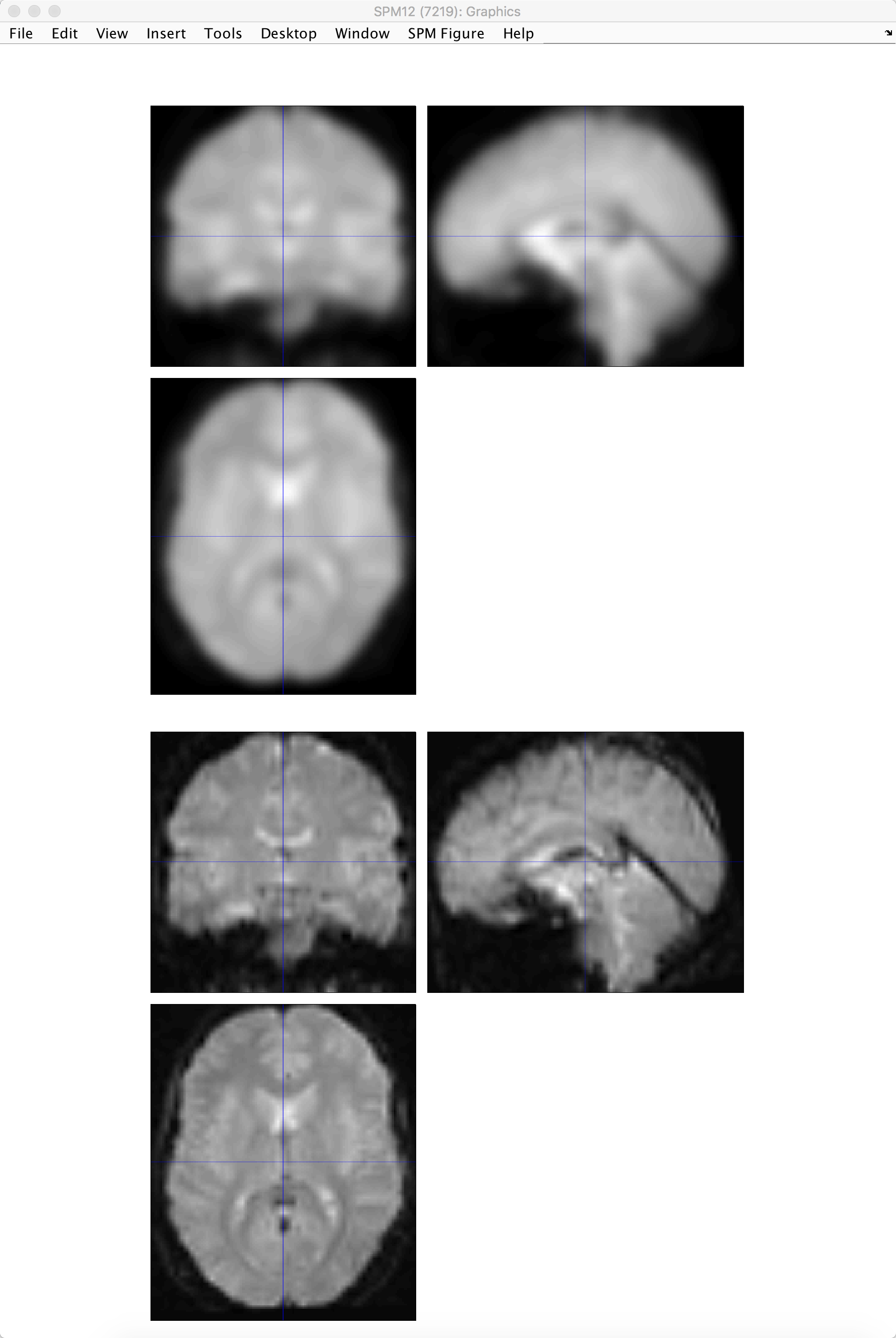
Checking the Smoothed Images
As before, use the Check Reg button to load a representative volume from the output you just created, and view it side by side with a warped functional image that hasn’t been smoothed. Does it look like the image has been smoothed the amount that you wanted?
Exercises
Try smoothing kernels of these different sizes (to save time, select just one volume from your preprocessed functional data):
0 0 0
3 3 3
30 30 30
Again, making sure to give each one a distinct Filename Prefix.
Before you look at the output, predict what it will look like. Does the result match your predictions? Include a screenshot of the output from each smoothing kernel.
Try smoothing in only one direction, e.g., by supplying a triplet of
[0 0 10]- which will smooth by 10mm in the z-direction. What do you notice about the results?Certain types of analyses, such as MVPA, require the time-series in each voxel to be as distinct as possible, in order to accurately classify a voxel as belonging to one condition or another. Would you predict that smoothing would be: (a) included in the preprocessing pipeline with the same smoothing kernel as a typical fMRI study; (b) not included in the preprocessing pipeline; (c) included with a reduced smoothing kernel compared to typical fMRI studies; or (d) included with an increased smoothing kernel compared to typical fMRI studies? Give reasons for your response.
Raw fMRI data already has some smoothness inherent in the data, even before doing any preprocessing. In your own words, describe what this is, and why you would see this in fMRI data. (Hint: If you know the time-series of one voxel, could you make an educated guess about what the time-series looks like in the neighboring voxels?) This will become an important topic to understand when we discuss cluster correction, which will be addressed in later chapters.
The smoothing kernel we apply here will add that smoothing size on top of the inherent smoothness already in the data. For example, if the inherent smoothness is 3mm, and we use a kernel of 8mm, the resulting smoothness after preprocessing will be around 11mm. Take a look at the AFNI command 3dBlurToFMWH, even if you don’t have AFNI installed. Read through the description and the recommendations. Would you prefer to use this command instead? Why or why not? As a reminder, some researchers prefer to integrate commands from several different software packages, depending on their needs; there is nothing invalid about substituting one package’s command for another’s, and then running the rest of the pipeline as usual.